Oncology NEWS International
- Oncology NEWS International Vol 16 No 6
- Volume 16
- Issue 6
Gene Expression Profiles of HCC Revealed on CT Scans
Researchers at Stanford and the University of California, San Diego (UCSD) have decoded the gene expression profiles of hepatocellular carcinomas (HCCs) noninvasively using the radiographic features of the tumors as seen on three-phase contrast-enhanced CT scans
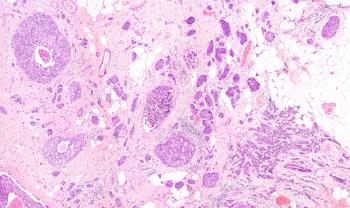
STANFORD, CaliforniaResearchers at Stanford and the University of California, San Diego (UCSD) have decoded the gene expression profiles of hepatocellular carcinomas (HCCs) noninvasively using the radiographic features of the tumors as seen on three-phase contrast-enhanced CT scans (see image on page 1). They also showed that imaging traits can be used to predict disease outcome, in the same way that gene assays such as OncotypeDx and MammaPrint predict breast cancer prognosis (Segal E et al: Nature Biotechnology, advance online publication May 21, 2007, DOI: 10.1038/nbt1307).
"Potentially in the future, one could use imaging to directly reveal multiple features of diseases that will make it much easier to carry out personalized medicine," said co-senior author Howard Chang, MD, PhD, assistant professor of dermatology at Stanford, who led the genomics arm of the study.
Michael Kuo, MD, assistant professor of interventional radiology, University of California, San Diego, and co-senior author, said that the work will help physicians obtain the molecular details of a specific tumor without having to remove body tissue for biopsy. Since tissue is not destroyed, the new technique allows for serial testing of the same tumor tissue.
Dr. Kuo began the research project in 2001 at Stanford. "Radiology seemed to be largely oblivious to the fundamental shift toward genomic, personalized medicine," he said in a news release. "Being there at Stanford [where microarray technology was first developed by Patrick Brown, MD, PhD], I was aware of that shift, and I was trying to think of ways to merge and integrate radiographic and genomic data." To increase the team's expertise in genomics and computational biology, in 2004 Dr. Kuo brought in Dr. Chang and lead author Eran Segal, PhD, currently with the Weizmann Institute of Science.
"When we look at noninvasive images, there are many different patterns that have no known meaning," Dr. Chang said. "Because imaging traits of tissues reflect the dynamic and physiologic interplay of parenchymal cells, blood vessels, and stroma, we reasoned that imaging traits may be used to predict gene expression patterns in human cancers."
Using 28 human HCC samples, the researchers defined and quantified 138 distinctive imaging traits present in one or more HCCs; they filtered the traits based on frequency and prominence to identify the 32 most informative traits. "For instance, many tumors displayed channels of radiodense signal on the arterial phase of the CT scan, and this trait was termed 'Internal Arteries,'" Dr. Chang said. They then used algorithms to match those imaging traits with a vast stockpile of microarray data generated from human HCC samples. They found that combinations of 28 imaging traits were sufficient to reconstruct about 80% of the global gene expression profiles of the tumors. Further, two predominant imaging traits (presence of "Internal Arteries" and absence of "Hypodense Halos") predicted microscopic venous invasion on histologic analysis in 30 HCC patients. "Internal Arteries" in the preoperative CT scan was a significant univariate predictor of overall survival in these patients and in another independent set of 32 patients.
v
Articles in this issue
over 18 years ago
ACS Launches Major Epidemiology Studyover 18 years ago
Pt Selection Key to Radioembolization of Liver Ca'sover 18 years ago
Million Dollar Gotham Prize Announcedover 18 years ago
Diagnostic Dilemma: GI Diseaseover 18 years ago
Junovan Fails to Win ODAC Nod for Osteosarcoma Treatmentover 18 years ago
Surveillance Colonoscopy Guidelines Not Being Followedover 18 years ago
Removing Stage IV Primary May Cut Mortalityover 18 years ago
Nuclear Export Inhibitors Testing Moving Forwardover 18 years ago
ODAC: orBec Yields No 'Substantial Efficacy' in GI GVHDNewsletter
Stay up to date on recent advances in the multidisciplinary approach to cancer.